728x90
반응형
import pandas as pd
url = "http://archive.ics.uci.edu/ml/machine-learning-databases/wine-quality/"
red = pd.read_csv(url+'winequality-red.csv',sep=';')
white = pd.read_csv(url+'winequality-white.csv',sep=';')
print(red.head())
fixed acidity volatile acidity citric acid residual sugar chlorides \
0 7.4 0.70 0.00 1.9 0.076
1 7.8 0.88 0.00 2.6 0.098
2 7.8 0.76 0.04 2.3 0.092
3 11.2 0.28 0.56 1.9 0.075
4 7.4 0.70 0.00 1.9 0.076
free sulfur dioxide total sulfur dioxide density pH sulphates \
0 11.0 34.0 0.9978 3.51 0.56
1 25.0 67.0 0.9968 3.20 0.68
2 15.0 54.0 0.9970 3.26 0.65
3 17.0 60.0 0.9980 3.16 0.58
4 11.0 34.0 0.9978 3.51 0.56
alcohol quality
0 9.4 5
1 9.8 5
2 9.8 5
3 9.8 6
4 9.4 5
red.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 1599 entries, 0 to 1598
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 fixed acidity 1599 non-null float64
1 volatile acidity 1599 non-null float64
2 citric acid 1599 non-null float64
3 residual sugar 1599 non-null float64
4 chlorides 1599 non-null float64
5 free sulfur dioxide 1599 non-null float64
6 total sulfur dioxide 1599 non-null float64
7 density 1599 non-null float64
8 pH 1599 non-null float64
9 sulphates 1599 non-null float64
10 alcohol 1599 non-null float64
11 quality 1599 non-null int64
dtypes: float64(11), int64(1)
memory usage: 150.0 KB
print(white.head())
fixed acidity volatile acidity citric acid residual sugar chlorides \
0 7.0 0.27 0.36 20.7 0.045
1 6.3 0.30 0.34 1.6 0.049
2 8.1 0.28 0.40 6.9 0.050
3 7.2 0.23 0.32 8.5 0.058
4 7.2 0.23 0.32 8.5 0.058
free sulfur dioxide total sulfur dioxide density pH sulphates \
0 45.0 170.0 1.0010 3.00 0.45
1 14.0 132.0 0.9940 3.30 0.49
2 30.0 97.0 0.9951 3.26 0.44
3 47.0 186.0 0.9956 3.19 0.40
4 47.0 186.0 0.9956 3.19 0.40
alcohol quality
0 8.8 6
1 9.5 6
2 10.1 6
3 9.9 6
4 9.9 6
white.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 4898 entries, 0 to 4897
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 fixed acidity 4898 non-null float64
1 volatile acidity 4898 non-null float64
2 citric acid 4898 non-null float64
3 residual sugar 4898 non-null float64
4 chlorides 4898 non-null float64
5 free sulfur dioxide 4898 non-null float64
6 total sulfur dioxide 4898 non-null float64
7 density 4898 non-null float64
8 pH 4898 non-null float64
9 sulphates 4898 non-null float64
10 alcohol 4898 non-null float64
11 quality 4898 non-null int64
dtypes: float64(11), int64(1)
memory usage: 459.3 KB
red['type'] = 0
white['type'] = 1
wine = pd.concat([red, white])
wine.info()
<class 'pandas.core.frame.DataFrame'>
Int64Index: 6497 entries, 0 to 4897
Data columns (total 13 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 fixed acidity 6497 non-null float64
1 volatile acidity 6497 non-null float64
2 citric acid 6497 non-null float64
3 residual sugar 6497 non-null float64
4 chlorides 6497 non-null float64
5 free sulfur dioxide 6497 non-null float64
6 total sulfur dioxide 6497 non-null float64
7 density 6497 non-null float64
8 pH 6497 non-null float64
9 sulphates 6497 non-null float64
10 alcohol 6497 non-null float64
11 quality 6497 non-null int64
12 type 6497 non-null int64
dtypes: float64(11), int64(2)
memory usage: 710.6 KB
# 데이터 정보
wine.describe()
fixed acidity volatile acidity citric acid residual sugar chlorides free sulfur dioxide total sulfur dioxide density pH sulphates alcohol quality type
count 6497.000000 6497.000000 6497.000000 6497.000000 6497.000000 6497.000000 6497.000000 6497.000000 6497.000000 6497.000000 6497.000000 6497.000000 6497.000000
mean 7.215307 0.339666 0.318633 5.443235 0.056034 30.525319 115.744574 0.994697 3.218501 0.531268 10.491801 5.818378 0.753886
std 1.296434 0.164636 0.145318 4.757804 0.035034 17.749400 56.521855 0.002999 0.160787 0.148806 1.192712 0.873255 0.430779
min 3.800000 0.080000 0.000000 0.600000 0.009000 1.000000 6.000000 0.987110 2.720000 0.220000 8.000000 3.000000 0.000000
25% 6.400000 0.230000 0.250000 1.800000 0.038000 17.000000 77.000000 0.992340 3.110000 0.430000 9.500000 5.000000 1.000000
50% 7.000000 0.290000 0.310000 3.000000 0.047000 29.000000 118.000000 0.994890 3.210000 0.510000 10.300000 6.000000 1.000000
75% 7.700000 0.400000 0.390000 8.100000 0.065000 41.000000 156.000000 0.996990 3.320000 0.600000 11.300000 6.000000 1.000000
max 15.900000 1.580000 1.660000 65.800000 0.611000 289.000000 440.000000 1.038980 4.010000 2.000000 14.900000 9.000000 1.000000
wine.type.value_counts()
1 4898
0 1599
Name: type, dtype: int64
wine_norm = (wine - wine.min()) / (wine.max() - wine.min())
wine_norm.describe()
fixed acidity volatile acidity citric acid residual sugar chlorides free sulfur dioxide total sulfur dioxide density pH sulphates alcohol quality type
count 6497.000000 6497.000000 6497.000000 6497.000000 6497.000000 6497.000000 6497.000000 6497.000000 6497.000000 6497.000000 6497.000000 6497.000000 6497.000000
mean 0.282257 0.173111 0.191948 0.074283 0.078129 0.102518 0.252868 0.146262 0.386435 0.174870 0.361131 0.469730 0.753886
std 0.107143 0.109758 0.087541 0.072972 0.058195 0.061630 0.130235 0.057811 0.124641 0.083599 0.172857 0.145543 0.430779
min 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000
25% 0.214876 0.100000 0.150602 0.018405 0.048173 0.055556 0.163594 0.100829 0.302326 0.117978 0.217391 0.333333 1.000000
50% 0.264463 0.140000 0.186747 0.036810 0.063123 0.097222 0.258065 0.149990 0.379845 0.162921 0.333333 0.500000 1.000000
75% 0.322314 0.213333 0.234940 0.115031 0.093023 0.138889 0.345622 0.190476 0.465116 0.213483 0.478261 0.500000 1.000000
max 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000 1.000000
wine_norm.head()
fixed acidity volatile acidity citric acid residual sugar chlorides free sulfur dioxide total sulfur dioxide density pH sulphates alcohol quality type
0 0.297521 0.413333 0.000000 0.019939 0.111296 0.034722 0.064516 0.206092 0.612403 0.191011 0.202899 0.333333 0.0
1 0.330579 0.533333 0.000000 0.030675 0.147841 0.083333 0.140553 0.186813 0.372093 0.258427 0.260870 0.333333 0.0
2 0.330579 0.453333 0.024096 0.026074 0.137874 0.048611 0.110599 0.190669 0.418605 0.241573 0.260870 0.333333 0.0
3 0.611570 0.133333 0.337349 0.019939 0.109635 0.055556 0.124424 0.209948 0.341085 0.202247 0.260870 0.500000 0.0
4 0.297521 0.413333 0.000000 0.019939 0.111296 0.034722 0.064516 0.206092 0.612403 0.191011 0.202899 0.333333 0.0
# wine_norm # 섞기
import numpy as np
wine_shuffle = wine_norm.sample(frac=1)
print(wine_shuffle.head())
fixed acidity volatile acidity citric acid residual sugar chlorides \
2593 0.206612 0.086667 0.174699 0.170245 0.064784
1273 0.305785 0.333333 0.120482 0.021472 0.106312
1348 0.280992 0.383333 0.018072 0.018405 0.114618
665 0.330579 0.133333 0.132530 0.012270 0.078073
1040 0.363636 0.400000 0.180723 0.023006 0.063123
free sulfur dioxide total sulfur dioxide density pH \
2593 0.166667 0.324885 0.148641 0.387597
1273 0.114583 0.087558 0.150954 0.294574
1348 0.020833 0.013825 0.168884 0.480620
665 0.079861 0.285714 0.140544 0.434109
1040 0.055556 0.304147 0.152111 0.387597
sulphates alcohol quality type
2593 0.089888 0.405797 0.333333 1.0
1273 0.117978 0.188406 0.333333 0.0
1348 0.095506 0.217391 0.333333 0.0
665 0.146067 0.217391 0.333333 1.0
1040 0.275281 0.405797 0.166667 1.0
wine_np = wine_shuffle.to_numpy()
print(wine_np[:5])
[[0.20661157 0.08666667 0.1746988 0.1702454 0.06478405 0.16666667
0.32488479 0.14864083 0.3875969 0.08988764 0.4057971 0.33333333
1. ]
[0.30578512 0.33333333 0.12048193 0.02147239 0.10631229 0.11458333
0.0875576 0.15095431 0.29457364 0.11797753 0.1884058 0.33333333
0. ]
[0.28099174 0.38333333 0.01807229 0.01840491 0.11461794 0.02083333
0.01382488 0.16888375 0.48062016 0.09550562 0.2173913 0.33333333
0. ]
[0.33057851 0.13333333 0.13253012 0.01226994 0.07807309 0.07986111
0.28571429 0.14054367 0.43410853 0.14606742 0.2173913 0.33333333
1. ]
[0.36363636 0.4 0.18072289 0.02300613 0.06312292 0.05555556
0.30414747 0.15211105 0.3875969 0.2752809 0.4057971 0.16666667
1. ]]
import tensorflow as tf
len(wine_np)
# 6497
train_idx = int(len(wine_np)*0.8)
print(train_idx)
train_X, train_Y = wine_np[:train_idx, : -1], wine_np[:train_idx, -1]
test_X, test_Y = wine_np[train_idx:, : -1], wine_np[train_idx:, -1]
# 5197
print(train_X[0])
print(train_Y[0])
print(test_X[0])
print(test_Y[0])
[0.15702479 0.09333333 0.12048193 0.23619632 0.05813953 0.13888889
0.24654378 0.22190091 0.3875969 0.13483146 0.13043478 0.5 ]
1.0
[0.23966942 0.10666667 0.21084337 0.19171779 0.06810631 0.21875
0.45852535 0.19066898 0.33333333 0.15730337 0.2173913 0.33333333]
1.0
# 원핫인코딩
import tensorflow as tf
train_Y = tf.keras.utils.to_categorical(train_Y, num_classes=2)
test_Y = tf.keras.utils.to_categorical(test_Y, num_classes=2)
train_Y[0]
# array([0., 1.], dtype=float32)
# 분류 모델
model = tf.keras.Sequential([tf.keras.layers.Dense(units=48, activation = 'relu', input_shape=(12,)),
tf.keras.layers.Dense(units=24, activation = 'relu'),
tf.keras.layers.Dense(units=12, activation = 'relu'),
tf.keras.layers.Dense(units=2, activation = 'softmax'),
tf.keras.layers.Dense(units=2, activation = 'sigmoid')
])
model.compile(optimizer=tf.keras.optimizers.Adam(lr=0.07),
# loss='categorical_crossentropy', metrics = ['accuracy'])
loss='binary_crossentropy', metrics = ['accuracy'])
model.summary()
# Dense 완전연결층, 1차원배열의 형태로 데이터 학습, 이미지의 픽셀의 관계 고려 안함
# 2차원 형태의 이미지를 1차원으로 변환, 이미지 특성 잃어버림
# 대신 convolution conv2D층
Model: "sequential"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
dense (Dense) (None, 48) 624
_________________________________________________________________
dense_1 (Dense) (None, 24) 1176
_________________________________________________________________
dense_2 (Dense) (None, 12) 300
_________________________________________________________________
dense_3 (Dense) (None, 2) 26
_________________________________________________________________
dense_4 (Dense) (None, 2) 6
=================================================================
Total params: 2,132
Trainable params: 2,132
Non-trainable params: 0
_________________________________________________________________
# 학습
history = model.fit(train_X, train_Y, epochs = 25, batch_size = 32, validation_split=0.25)
Epoch 1/25
122/122 [==============================] - 2s 8ms/step - loss: 0.5704 - accuracy: 0.7708 - val_loss: 0.5771 - val_accuracy: 0.7362
Epoch 2/25
122/122 [==============================] - 0s 4ms/step - loss: 0.5489 - accuracy: 0.7645 - val_loss: 0.5807 - val_accuracy: 0.7362
Epoch 3/25
122/122 [==============================] - 0s 4ms/step - loss: 0.5497 - accuracy: 0.7649 - val_loss: 0.5775 - val_accuracy: 0.7362
Epoch 4/25
122/122 [==============================] - 0s 4ms/step - loss: 0.5431 - accuracy: 0.7682 - val_loss: 0.5825 - val_accuracy: 0.7362
Epoch 5/25
122/122 [==============================] - 0s 4ms/step - loss: 0.5570 - accuracy: 0.7586 - val_loss: 0.5770 - val_accuracy: 0.7362
Epoch 6/25
122/122 [==============================] - 0s 4ms/step - loss: 0.5663 - accuracy: 0.7477 - val_loss: 0.5775 - val_accuracy: 0.7362
Epoch 7/25
122/122 [==============================] - 0s 4ms/step - loss: 0.5632 - accuracy: 0.7504 - val_loss: 0.5870 - val_accuracy: 0.7362
Epoch 8/25
122/122 [==============================] - 0s 4ms/step - loss: 0.5582 - accuracy: 0.7556 - val_loss: 0.5773 - val_accuracy: 0.7362
Epoch 9/25
122/122 [==============================] - 0s 4ms/step - loss: 0.5576 - accuracy: 0.7564 - val_loss: 0.5783 - val_accuracy: 0.7362
Epoch 10/25
122/122 [==============================] - 0s 4ms/step - loss: 0.5713 - accuracy: 0.7444 - val_loss: 0.5772 - val_accuracy: 0.7362
Epoch 11/25
122/122 [==============================] - 0s 4ms/step - loss: 0.5606 - accuracy: 0.7534 - val_loss: 0.5799 - val_accuracy: 0.7362
Epoch 12/25
122/122 [==============================] - 0s 4ms/step - loss: 0.5498 - accuracy: 0.7637 - val_loss: 0.5787 - val_accuracy: 0.7362
Epoch 13/25
122/122 [==============================] - 0s 4ms/step - loss: 0.5517 - accuracy: 0.7620 - val_loss: 0.5854 - val_accuracy: 0.7362
Epoch 14/25
122/122 [==============================] - 0s 4ms/step - loss: 0.5663 - accuracy: 0.7488 - val_loss: 0.5780 - val_accuracy: 0.7362
Epoch 15/25
122/122 [==============================] - 0s 4ms/step - loss: 0.5583 - accuracy: 0.7560 - val_loss: 0.5772 - val_accuracy: 0.7362
Epoch 16/25
122/122 [==============================] - 0s 4ms/step - loss: 0.5645 - accuracy: 0.7501 - val_loss: 0.5770 - val_accuracy: 0.7362
Epoch 17/25
122/122 [==============================] - 0s 4ms/step - loss: 0.5686 - accuracy: 0.7456 - val_loss: 0.5860 - val_accuracy: 0.7362
Epoch 18/25
122/122 [==============================] - 0s 4ms/step - loss: 0.5499 - accuracy: 0.7620 - val_loss: 0.5788 - val_accuracy: 0.7362
Epoch 19/25
122/122 [==============================] - 0s 4ms/step - loss: 0.5738 - accuracy: 0.7478 - val_loss: 0.5802 - val_accuracy: 0.7362
Epoch 20/25
122/122 [==============================] - 0s 4ms/step - loss: 0.5537 - accuracy: 0.7598 - val_loss: 0.5800 - val_accuracy: 0.7362
Epoch 21/25
122/122 [==============================] - 0s 4ms/step - loss: 0.5449 - accuracy: 0.7661 - val_loss: 0.5911 - val_accuracy: 0.7362
Epoch 22/25
122/122 [==============================] - 0s 4ms/step - loss: 0.5615 - accuracy: 0.7542 - val_loss: 0.5809 - val_accuracy: 0.7362
Epoch 23/25
122/122 [==============================] - 0s 4ms/step - loss: 0.5647 - accuracy: 0.7507 - val_loss: 0.5771 - val_accuracy: 0.7362
Epoch 24/25
122/122 [==============================] - 0s 4ms/step - loss: 0.5590 - accuracy: 0.7544 - val_loss: 0.5774 - val_accuracy: 0.7362
Epoch 25/25
122/122 [==============================] - 0s 4ms/step - loss: 0.5519 - accuracy: 0.7623 - val_loss: 0.5882 - val_accuracy: 0.7362
# 결과 시각화 loss acc
import matplotlib.pyplot as plt
plt.figure(figsize = (12,4))
plt.subplot(1,2,1)
plt.plot(history.history['loss'], 'b-', label = 'loss')
plt.plot(history.history['val_loss'], 'r--', label = 'val_loss')
plt.xlabel('Epoch')
plt.legend()
plt.show()
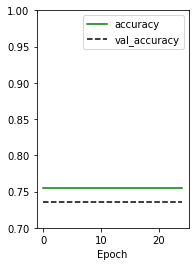
plt.subplot(1,2,2)
plt.plot(history.history['accuracy'], 'g-', label = 'accuracy')
plt.plot(history.history['val_accuracy'], 'k--', label = 'val_accuracy')
plt.xlabel('Epoch')
plt.ylim(0.7, 1)
plt.legend()
plt.show()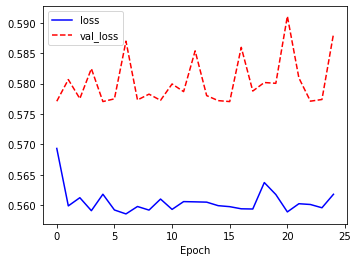
# 모델평가
model.evaluate(test_X, test_Y)
41/41 [==============================] - 0s 2ms/step - loss: 0.5416 - accuracy: 0.7700
[0.5416469573974609, 0.7699999809265137]
반응형
'Data_Science > Data_Analysis_Py' 카테고리의 다른 글
| 48. Fashion MNIST || convolution (0) | 2021.11.26 |
|---|---|
| 47. Cat image, convolution 설명 (0) | 2021.11.25 |
| 45. 보스턴주택가격 딥러닝 회귀 예측 평가 (0) | 2021.11.25 |
| 44. 보스턴주택가격 회귀예측 || kfold (0) | 2021.11.25 |
| 43. 보스턴주택가격 딥러닝 회귀 예측 || hold-out (0) | 2021.11.25 |